blog
2022/04/28
Running Beam SQL in notebooksNing Kang [@ningkang0957]
Intro
Beam SQL allows a Beam user to query PCollections with SQL statements. Interactive Beam provides an integration between Apache Beam and Jupyter Notebooks (formerly known as IPython Notebooks) to make pipeline prototyping and data exploration much faster and easier. You can set up your own notebook user interface (for example, JupyterLab or classic Jupyter Notebooks) on your own device following their documentations. Alternatively, you can choose a hosted solution that does everything for you. You are free to select whichever notebook user interface you prefer. For simplicity, this post does not go through the notebook environment setup and uses Apache Beam Notebooks that provides a cloud-hosted JupyterLab environment and lets a Beam user iteratively develop pipelines, inspect pipeline graphs, and parse individual PCollections in a read-eval-print-loop (REPL) workflow.
In this post, you will see how to use beam_sql, a notebook
magic, to
execute Beam SQL in notebooks and inspect the results.
By the end of the post, it also demonstrates how to use the beam_sql magic
with a production environment, such as running it as a one-shot job on
Dataflow. It’s optional. To follow those steps, you should have a project in
Google Cloud Platform with
necessary APIs enabled
, and you should have enough permissions to create a Google Cloud Storage bucket
(or to use an existing one), query a public Google Cloud BigQuery dataset, and
run Dataflow jobs.
If you choose to use the cloud hosted notebook solution, once you have your Google Cloud project ready, you will need to create an Apache Beam Notebooks instance and open the JupyterLab web interface. Please follow the instructions given at: https://cloud.google.com/dataflow/docs/guides/interactive-pipeline-development#launching_an_notebooks_instance
Getting familiar with the environment
Landing page
After starting your own notebook user interface: for example, if using Apche
Beam Notebooks, after clicking the OPEN JUPYTERLAB link, you will land on
the default launcher page of the notebook environment.
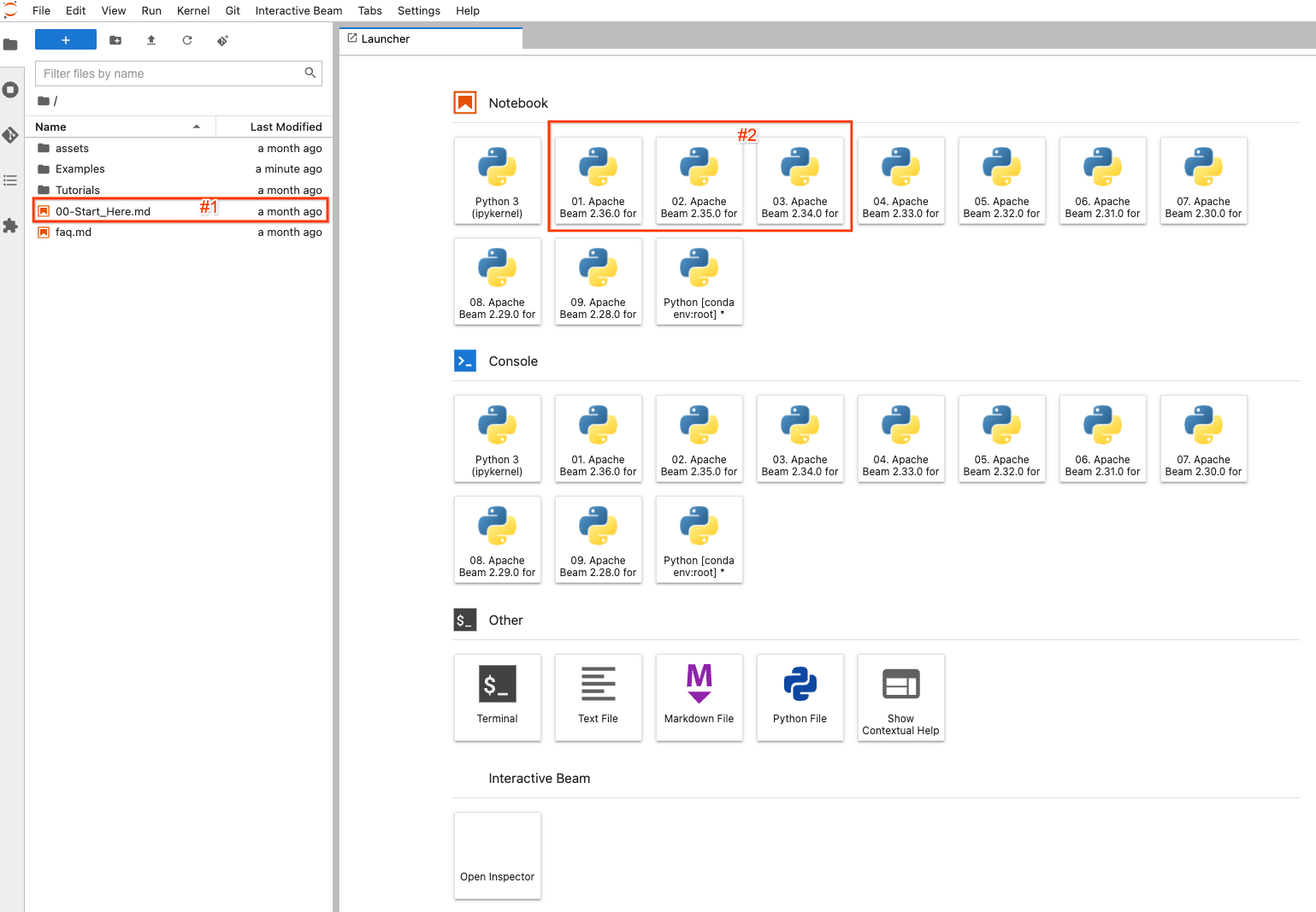
On the left side, there is a file explorer to view examples, tutorials and
assets on the notebook instance. To easily navigate the files, you may
double-click the 00-Start_Here.md (#1 in the screenshot) file to view detailed
information about the files.
On the right side, it displays the default launcher page of JupyterLab. To
create and open a completely new notebook file and code with a selected version
of Apache Beam, click one of (#2) the items with Apache Beam >=2.34.0 (because
beam_sql was introduced in 2.34.0) installed.
Create/open a notebook
For example, if you clicked the image button with Apache Beam 2.36.0, you would
see an Untitled.ipynb file created and opened.
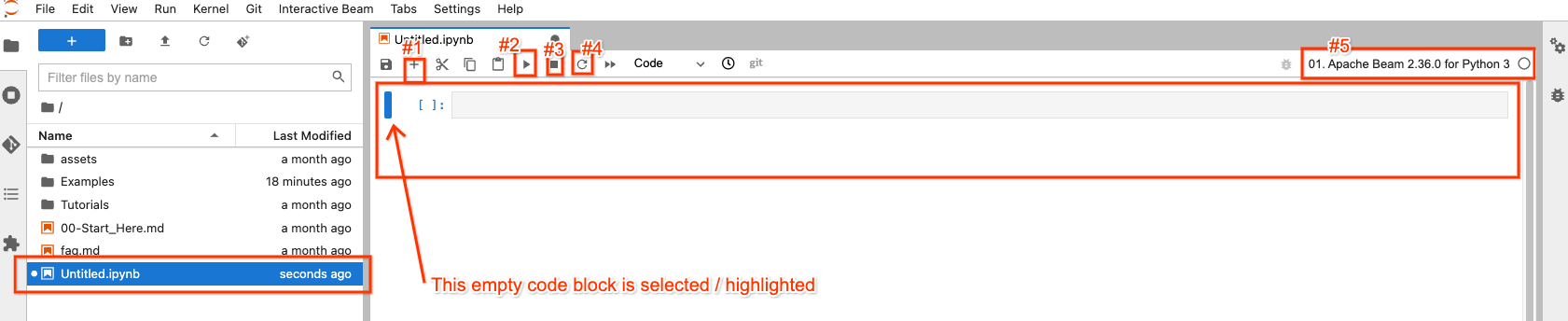
In the file explorer, your new notebook file has been created as
Untitled.ipynb.
On the right side, in the opened notebook, there are 4 buttons on top that you may interact most frequently with:
- #1: insert an empty code block after the selected / highlighted code block
- #2: execute the code in the block that is selected / highlighted
- #3: interrupt code execution if your code execution is stuck
- #4: “Restart the kernel”: clear all states from code executions and start from fresh
There is a button on the top-right (#5) for you to choose a different Apache Beam version if needed, so it’s not set in stone.
You can always double-click a file from the file explorer to open it without creating a new one.
Beam SQL
beam_sql magic
beam_sql is an IPython
custom magic.
If you’re not familiar with magics, here are some
built-in examples.
It’s a convenient way to validate your queries locally against known/test data
sources when prototyping a Beam pipeline with SQL, before productionizing it on
remote cluster/services.
The Apache Beam Notebooks environment has preloaded the beam_sql magic and
basic apache-beam modules so you can directly use them without additional
imports. You can also explicitly load the magic via
%load_ext apache_beam.runners.interactive.sql.beam_sql_magics and
apache-beam modules if you set up your own notebook elsewhere.
You can type:
%beam_sql -h
and then execute the code to learn how to use the magic:
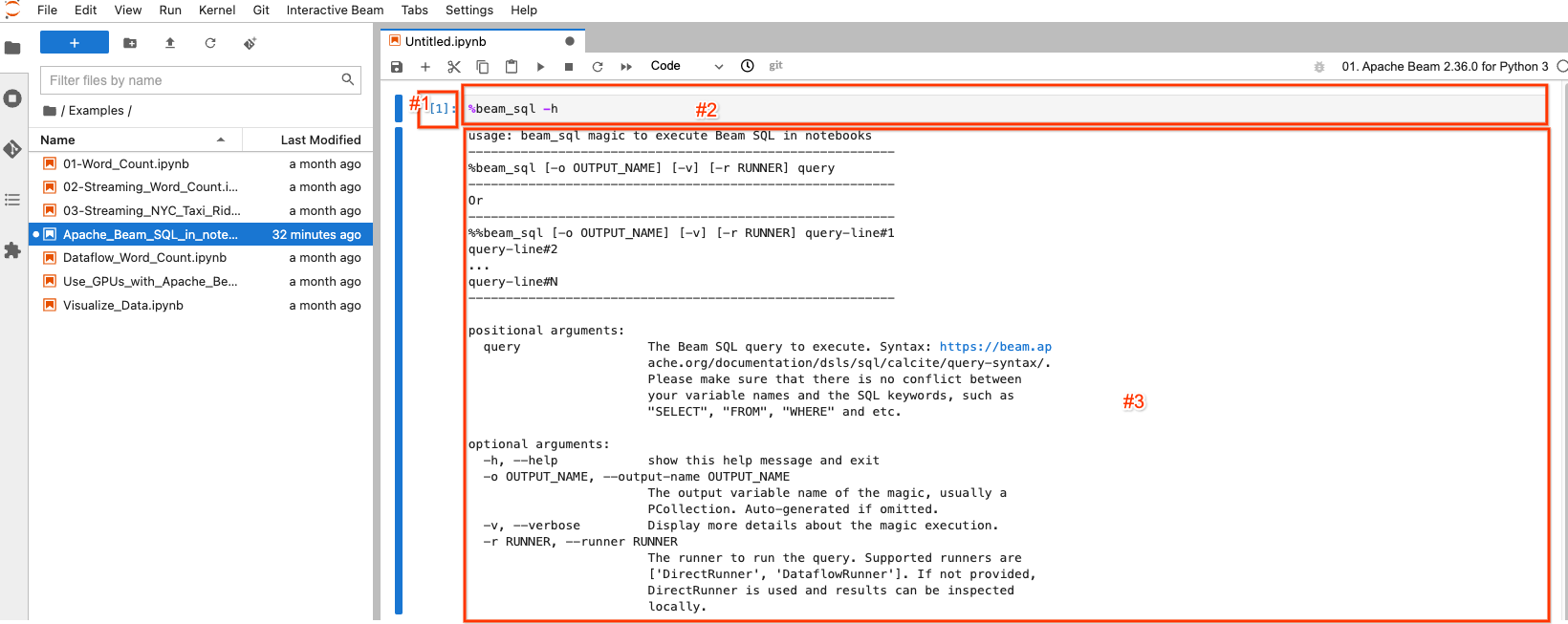
The selected/highlighted block is called a notebook cell. It mainly has 3 components:
- #1: The execution count.
[1]indicates this block is the first executed code. It increases by 1 for each piece of code you execute even if you re-execute the same piece of code.[ ]indicates this block is not executed. - #2: The cell input: the code gets executed.
- #3: The cell output: the output of the code execution. Here it contains the
help documentation of the
beam_sqlmagic.
Create a PCollection
There are 3 scenarios for Beam SQL when creating a PCollection:
- Use Beam SQL to create a PCollection from constant values
%%beam_sql -o pcoll
SELECT CAST(1 AS INT) AS id, CAST('foo' AS VARCHAR) AS str, CAST(3.14 AS DOUBLE) AS flt

The beam_sql magic creates and outputs a PCollection named pcoll with
element_type like BeamSchema_...(id: int32, str: str, flt: float64).
Note that you have not explicitly created a Beam pipeline. You get a
PCollection because the beam_sql magic always implicitly creates a pipeline to
execute your SQL query. To hold the elements with each field’s type info, Beam
automatically creates a
schema
as the element_type for the created PCollection. You will learn more about
schema-aware PCollections later.
- Use Beam SQL to query a PCollection
You can chain another SQL using the output from a previous SQL (or any schema-aware PCollection produced by any normal Beam PTransforms) as the input to produce a new PCollection.
Note: if you name the output PCollection, make sure that it’s unique in your notebook to avoid overwriting a different PCollection.
%%beam_sql -o id_pcoll
SELECT id FROM pcoll

- Use Beam SQL to join multiple PCollections
You can query multiple PCollections from a single query.
%%beam_sql -o str_with_same_id
SELECT id, str FROM pcoll JOIN id_pcoll USING (id)

Now you have learned how to use the beam_sql magic to create PCollections and
inspect their results.
Tip: if you accidentally delete some of the notebook cell outputs, you can
always check the content of a PCollection by invoking ib.show(pcoll_name) or
ib.collect(pcoll_name) where ib stands for “Interactive Beam”
(learn more).
Schema-aware PCollections
The beam_sql magic provides the flexibility to seamlessly mix SQL and non-SQL
Beam statements to build pipelines and even run them on Dataflow. However, each
PCollection queried by Beam SQL needs to have a
schema.
For the beam_sql magic, it’s recommended to use typing.NamedTuple when a
schema is desired. You can go through the below example to learn more details
about schema-aware PCollections.
Setup
In the setup of this example, you will:
- Install PyPI package
namesusing the built-in%pipmagic: you will use the module to generate some random English names as the raw data input. - Define a schema with
NamedTuplethat has 2 attributes:id- an unique numeric identifier of a person;name- a string name of a person. - Define a pipeline with an
InteractiveRunnerto utilize notebook related features of Apache Beam.
%pip install names
import names
from typing import NamedTuple
class Person(NamedTuple):
id: int
name: str
p = beam.Pipeline(InteractiveRunner())
There is no visible output for the code execution.
Create schema-aware PCollections without using SQL
persons = (p
| beam.Create([Person(id=x, name=names.get_full_name()) for x in range(10)]))
ib.show(persons)

persons_2 = (p
| beam.Create([Person(id=x, name=names.get_full_name()) for x in range(5, 15)]))
ib.show(persons_2)

Now you have 2 PCollections both with the same schema defined by the Person
class:
personscontains 10 records for 10 persons with ids ranging from 0 to 9,persons_2contains another 10 records for 10 persons with ids ranging from 5 to 14.
Encode and Decode of schema-aware PCollections
For this example, you still need one more piece of data from the first pcoll
that you have created with instructions in this post.
You can use the original pcoll. Optionally, if you want to exercise using
coders explicitly with schema-aware PCollections, you can add a Text I/O into
the mix: write the content of pcoll into a text file retaining its schema
information, then read the file back into a new schema-aware PCollection called
pcoll_in_file, and use the new PCollection to join persons and persons_2
to find names with the common id in all three of them.
To encode pcoll into a file, execute:
coder=beam.coders.registry.get_coder(pcoll.element_type)
pcoll | beam.io.textio.WriteToText('/tmp/pcoll', coder=coder)
pcoll.pipeline.run().wait_until_finish()
!cat /tmp/pcoll*

The above code execution writes the PCollection pcoll (basically
{id: 1, str: foo, flt: 3.14}) into a text file using the coder assigned by
Beam. As you can see, the file content is recorded in a binary non
human-readable format, and that’s normal.
To decode the file content into a new PCollection, execute:
pcoll_in_file = p | beam.io.ReadFromText(
'/tmp/pcoll*', coder=coder).with_output_types(
pcoll.element_type)
ib.show(pcoll_in_file)

Note you have to use the same coder during encoding and decoding, and
furthermore you may assign the schema explicitly to the new PCollection through
with_output_types().
Reading out the encoded binary content from the text file and decoding it with
the correct coder, the content of pcoll is recovered into pcoll_in_file. You
can use this technique to save and share your data through any Beam I/O (not
necessarily a text file) with collaborators who work on their own pipelines (not
just in your notebook session or pipelines).
Schema in beam_sql magic
The beam_sql magic automatically registers a RowCoder for your NamedTuple
schema so that you only need to focus on preparing your data for query without
worrying about coders. To see more verbose details of what the beam_sql magic
does behind the scenes, you can use the -v option.
For example, you can look for all elements with id < 5 in persons with the
below query and assign the output to persons_id_lt_5.
%%beam_sql -o persons_id_lt_5 -v
SELECT * FROM persons WHERE id < 5
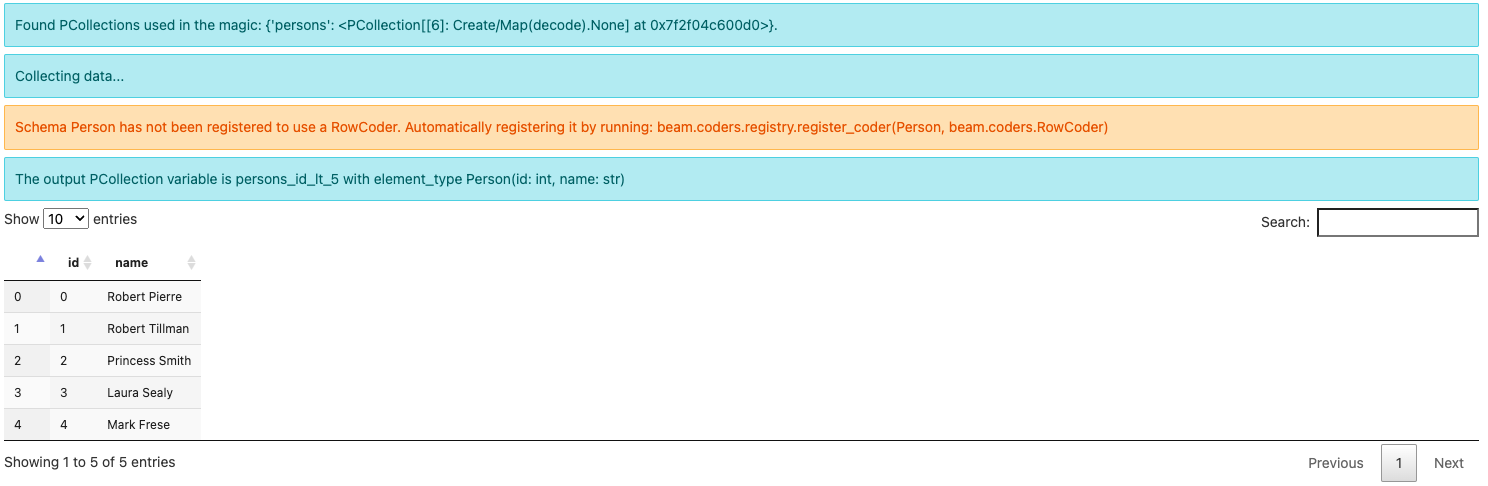
Since this is the first time running this query, you might see a warning message about:
Schema Person has not been registered to use a RowCoder. Automatically registering it by running: beam.coders.registry.register_coder(Person, beam.coders.RowCoder)
The beam_sql magic helps registering a RowCoder for each schema you define
and use whenever it finds one. You can also explicitly run the same code to do
so.
Note the output element type is Person(id: int, name: str) instead of
BeamSchema_… because you have selected all the fields from a single
PCollection of the known type Person(id: int, name: str).
Another example, you can query for all names from persons and persons_2 with
the same ids and assign the output to persons_with_common_id:
%%beam_sql -o persons_with_common_id -v
SELECT * FROM persons JOIN persons_2 USING (id)
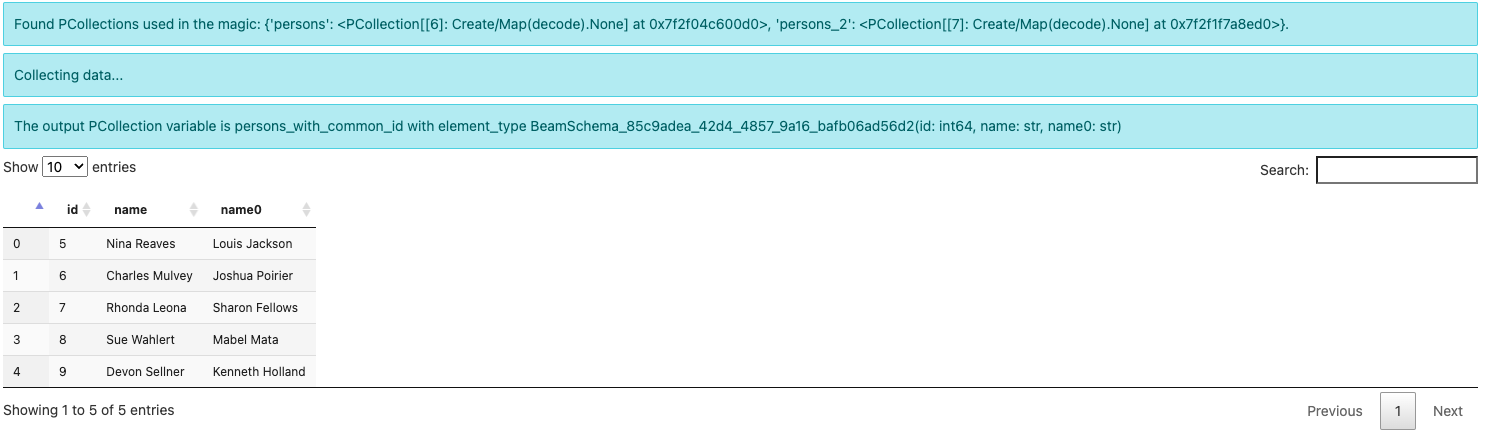
Note the output element type is now some
BeamSchema_...(id: int64, name: str, name0: str). Because you have selected
columns from both PCollections, there is no known schema to hold the result.
Beam automatically creates a schema and differentiates the conflicted field
name by suffixing 0 to one of them.
And since Person is already previously registered with a RowCoder, there is
no more warning about registering it even with the -v option.
Additionally, you can do a join with pcoll_in_file, persons and persons_2:
%%beam_sql -o entry_with_common_id
SELECT pcoll_in_file.id, persons.name AS name_1, persons_2.name AS name_2
FROM pcoll_in_file JOIN persons ON pcoll_in_file.id = persons.id
JOIN persons_2 ON pcoll_in_file.id = persons_2.id

The schema generated reflects the column renaming you have done in the SQL.
An Example
You will go through an example to find out the US state with the most COVID positive cases on a specific day with data provided by the covid tracking project.
Get the data
import json
import requests
# The covidtracking project has stopped collecting new data, current data ends on 2021-03-07
json_current='https://api.covidtracking.com/v1/states/current.json'
def get_json_data(url):
with requests.Session() as session:
data = json.loads(session.get(url).text)
return data
current_data = get_json_data(json_current)
current_data[0]

The data is dated as 2021-03-07. It contains many details about COVID cases for
different states in the US. current_data[0] is just one of the data points.
You can get rid of most of the columns of the data. For example, just focus on
“date”, “state”, “positive” and “negative”, and then define a schema
UsCovidData:
from typing import Optional
class UsCovidData(NamedTuple):
partition_date: str # Remember to str(e['date']).
state: str
positive: int
negative: Optional[int]
Note:
dateis a keyword in (Calcite)SQL, use a different field name such aspartition_date;datefrom the data is aninttype, notstr. Make sure you convert the data usingstr()or usedate: int.negativehas missing values and the default isNone. So instead ofnegative: int, it should benegative: Optional[int]. Or you can convertNoneinto 0 when using the schema.
Then parse the json data into a PCollection with the schema:
p_sql = beam.Pipeline(runner=InteractiveRunner())
covid_data = (p_sql
| 'Create PCollection from json' >> beam.Create(current_data)
| 'Parse' >> beam.Map(
lambda e: UsCovidData(
partition_date=str(e['date']),
state=e['state'],
positive=e['positive'],
negative=e['negative'])).with_output_types(UsCovidData))
ib.show(covid_data)

Query
You can now find the biggest positive on the “current day” (2021-03-07).
%%beam_sql -o max_positive
SELECT partition_date, MAX(positive) AS positive
FROM covid_data
GROUP BY partition_date

However, this is just the positive number. You cannot observe the state that has this maximum number nor the negative case number for the state.
To enrich your result, you have to join this data back to the original data set you have parsed.
%%beam_sql -o entry_with_max_positive
SELECT covid_data.partition_date, covid_data.state, covid_data.positive, {fn IFNULL(covid_data.negative, 0)} AS negative
FROM covid_data JOIN max_positive
USING (partition_date, positive)

Now you can see all columns of the data with the maximum positive case on
2021-03-07.
Note: to handle missing values of the negative column in the original data,
you can use {fn IFNULL(covid_data.negative, 0)} to set null values to 0.
When you’re ready to scale up, you can translate the SQLs into a pipeline with
SqlTransforms and run your pipeline on a distributed runner like Flink or
Spark. This post demonstrates it by launching a one-shot job on Dataflow from
the notebook with the help of beam_sql magic.
Run on Dataflow
Now that you have a pipeline that parses US COVID data from json to find positive/negative/state information for the state with the most positive cases on each day, you can try applying it to all historical daily data and running it on Dataflow.
The new data source you will use is a public dataset from USAFacts US Coronavirus Database that contains all historical daily summary of COVID cases in the US.
The schema of data is very similar to what the covid tracking project website
provides. The fields you will query are: date, state, confirmed_cases, and
deaths.
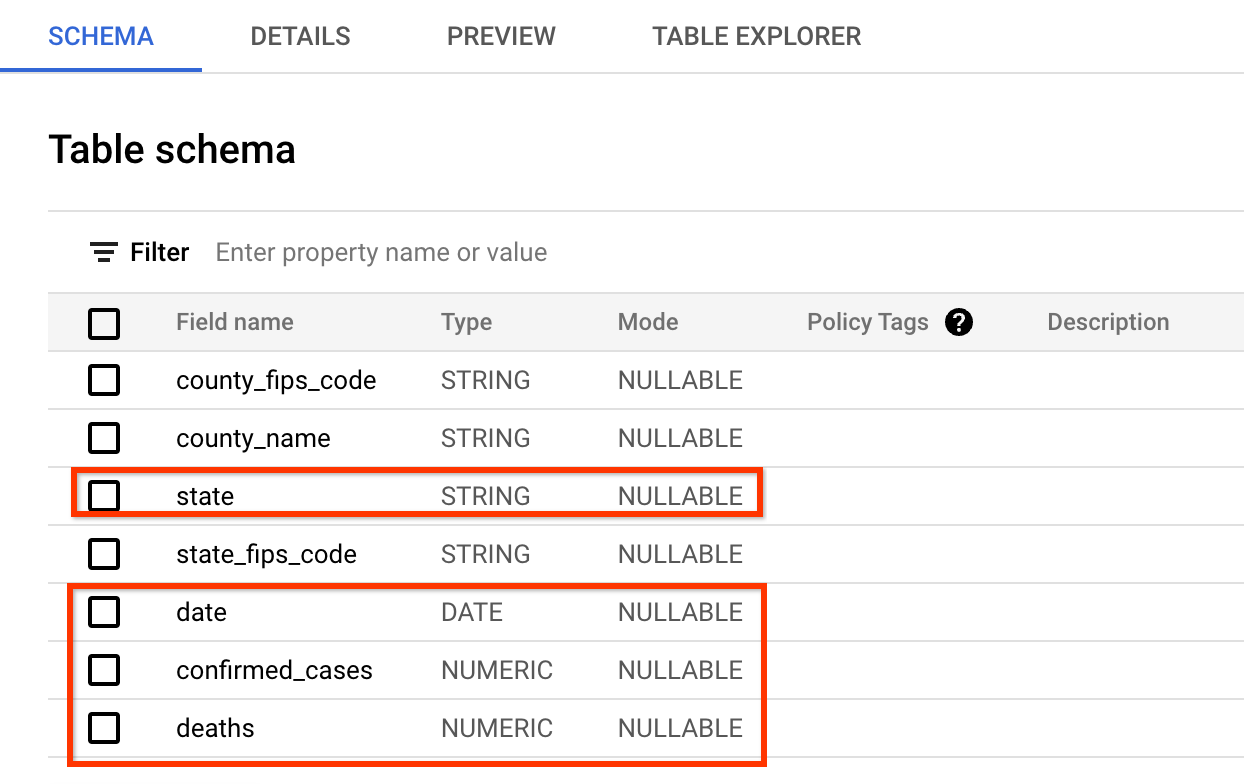
A preview of the data looks like below (you may skip the inspection in BigQuery and just take a look at the screenshot):
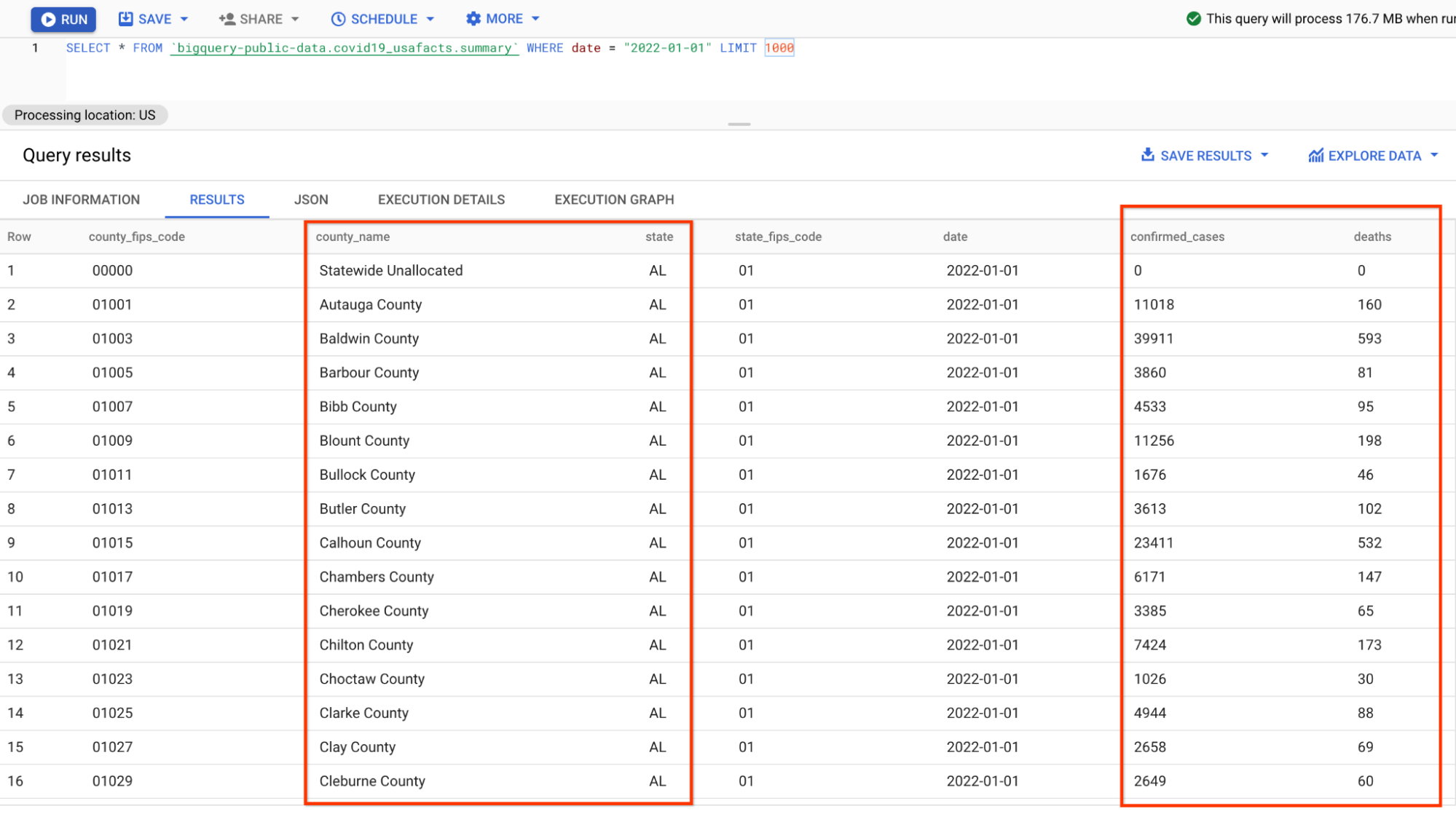
The format of the data is slightly different from the json data you parsed in the previous pipeline because the numbers are grouped by counties instead of states, thus some additional aggregations need to be done in the SQLs.
If you need a fresh execution, you may click the “Restart the kernel” button on the top menu.
Full code is as below, on-top of the original pipeline and queries:
- It changes the source from a single-day data to a more complete historical data;
- It changes the I/O and schema to accommodate the new dataset;
- It changes the SQLs to include more aggregations to accommodate the new format of the dataset.
Prepare the data with schema
from typing import NamedTuple
from typing import Optional
# Public BQ dataset.
table = 'bigquery-public-data:covid19_usafacts.summary'
# Replace with your project.
project = 'YOUR-PROJECT-NAME-HERE'
# Replace with your GCS bucket.
gcs_location = 'gs://YOUR_GCS_BUCKET_HERE'
class UsCovidData(NamedTuple):
partition_date: str
state: str
confirmed_cases: Optional[int]
deaths: Optional[int]
p_on_dataflow = beam.Pipeline(runner=InteractiveRunner())
covid_data = (p_on_dataflow
| 'Read dataset' >> beam.io.ReadFromBigQuery(
project=project, table=table, gcs_location=gcs_location)
| 'Parse' >> beam.Map(
lambda e: UsCovidData(
partition_date=str(e['date']),
state=e['state'],
confirmed_cases=int(e['confirmed_cases']),
deaths=int(e['deaths']))).with_output_types(UsCovidData))
Run on Dataflow
To run SQL on Dataflow is very simple, you just need to add the option
-r DataflowRunner.
%%beam_sql -o data_by_state -r DataflowRunner
SELECT partition_date, state, SUM(confirmed_cases) as confirmed_cases, SUM(deaths) as deaths
FROM covid_data
GROUP BY partition_date, state
Different from previous beam_sql magic executions, you won’t see the result
immediately. Instead, a form like below is printed in the notebook cell output:
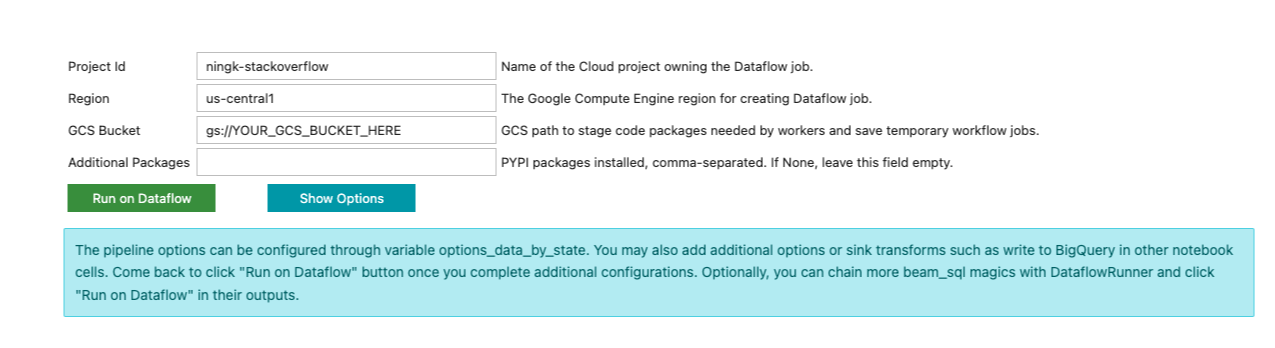
The beam_sql magic tries its best to guess your project id and preferred cloud
region. You still have to input additional information necessary to submit a
Dataflow job, such as a GCS bucket to stage the Dataflow job and any additional
Python dependencies the job needs.
For now, ignore the form in the cell output, because you still need 2 more SQLs
to: 1) find the maximum confirmed cases on each day; 2) join the maximum case
data with the full data_by_state. The beam_sql magic allows you to chain SQLs,
so chain 2 more by executing:
%%beam_sql -o max_cases -r DataflowRunner
SELECT partition_date, MAX(confirmed_cases) as confirmed_cases
FROM data_by_state
GROUP BY partition_date
And
%%beam_sql -o data_with_max_cases -r DataflowRunner
SELECT data_by_state.partition_date, data_by_state.state, data_by_state.confirmed_cases, data_by_state.deaths
FROM data_by_state JOIN max_cases
USING (partition_date, confirmed_cases)
By default, when running beam_sql on Dataflow, the output PCollection will be
written to a text file on GCS. The “write” is automatically provided by
beam_sql and mainly for your inspection of the output data for this one-shot
Dataflow job. It’s lightweight and does not encode elements for further
development. To save the output and share it with others, you can add more Beam
I/Os into the mix.
For example, you can appropriately encode elements into text files using the technique described in the above schema-aware PCollections example.
from apache_beam.options.pipeline_options import GoogleCloudOptions
coder = beam.coders.registry.get_coder(data_with_max_cases.element_type)
max_data_file = gcs_location + '/encoded_max_data'
data_with_max_cases | beam.io.textio.WriteToText(max_data_file, coder=coder)
Furthermore, you can create a new BQ dataset in your own project to store the processed data.
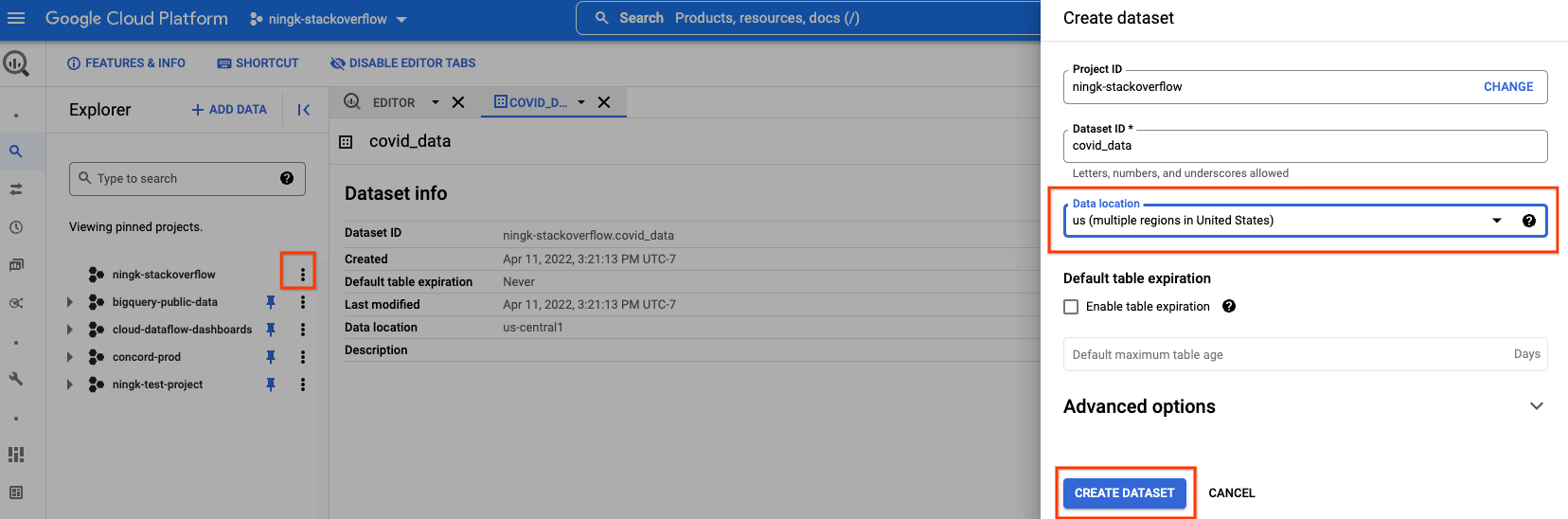
You have to select the same data location as the public BigQuery data you are reading. In this case, “us (multiple regions in United States)”.
Once you finish creating an empty dataset, you can execute below:
output_table=f'{project}:covid_data.max_analysis'
bq_schema = {
'fields': [
{'name': 'partition_date', 'type': 'STRING'},
{'name': 'state', 'type': 'STRING'},
{'name': 'confirmed_cases', 'type': 'INTEGER'},
{'name': 'deaths', 'type': 'INTEGER'}]}
(data_with_max_cases
| 'To json-like' >> beam.Map(lambda x: {
'partition_date': x.partition_date,
'state': x.state,
'confirmed_cases': x.confirmed_cases,
'deaths': x.deaths})
| beam.io.WriteToBigQuery(
table=output_table,
schema=bq_schema,
method='STREAMING_INSERTS',
custom_gcs_temp_location=gcs_location))
Now back in the form of the last SQL cell output, you may fill in necessary information to run the pipeline on Dataflow. An example input looks like below:
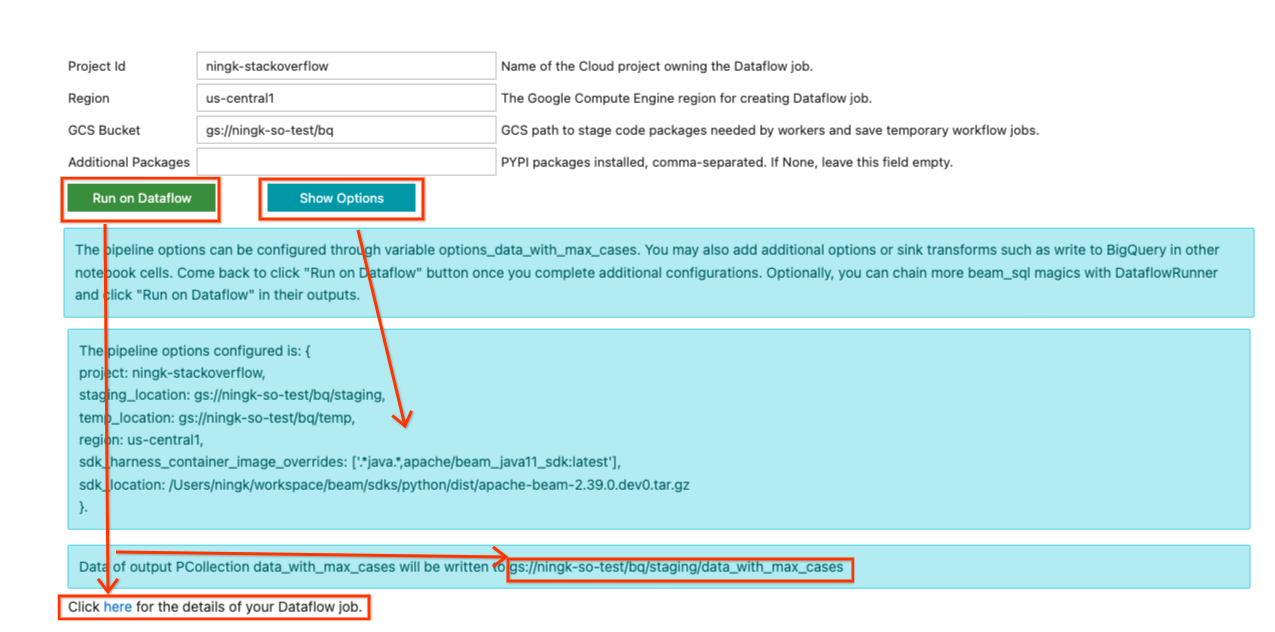
Because this pipeline doesn’t use any additional Python dependency, “Additional
Packages” is left empty. In the previous example where you have installed a
package called names, to run that pipeline on Dataflow, you have to put
names in this field.
Once you finish updating your inputs, you can click the Show Options button to
view what pipeline options have been configured based on your inputs. A variable
options_[YOUR_OUTPUT_PCOLL_NAME] is generated, and you can supply more
pipeline options to it if the form is not enough for your execution.
Once you are ready to submit the Dataflow job, click the Run on Dataflow
button. It tells you where the default output would be written, and after a
while, a line with:
Click here for the details of your Dataflow job.
would be displayed. You can click on the hyperlink to go to your Dataflow job page. (Optionally, you can ignore the form and continue development to extend your pipeline. Once you are satisfied with the state of your pipeline, you can come back to the form and submit the job to Dataflow.)
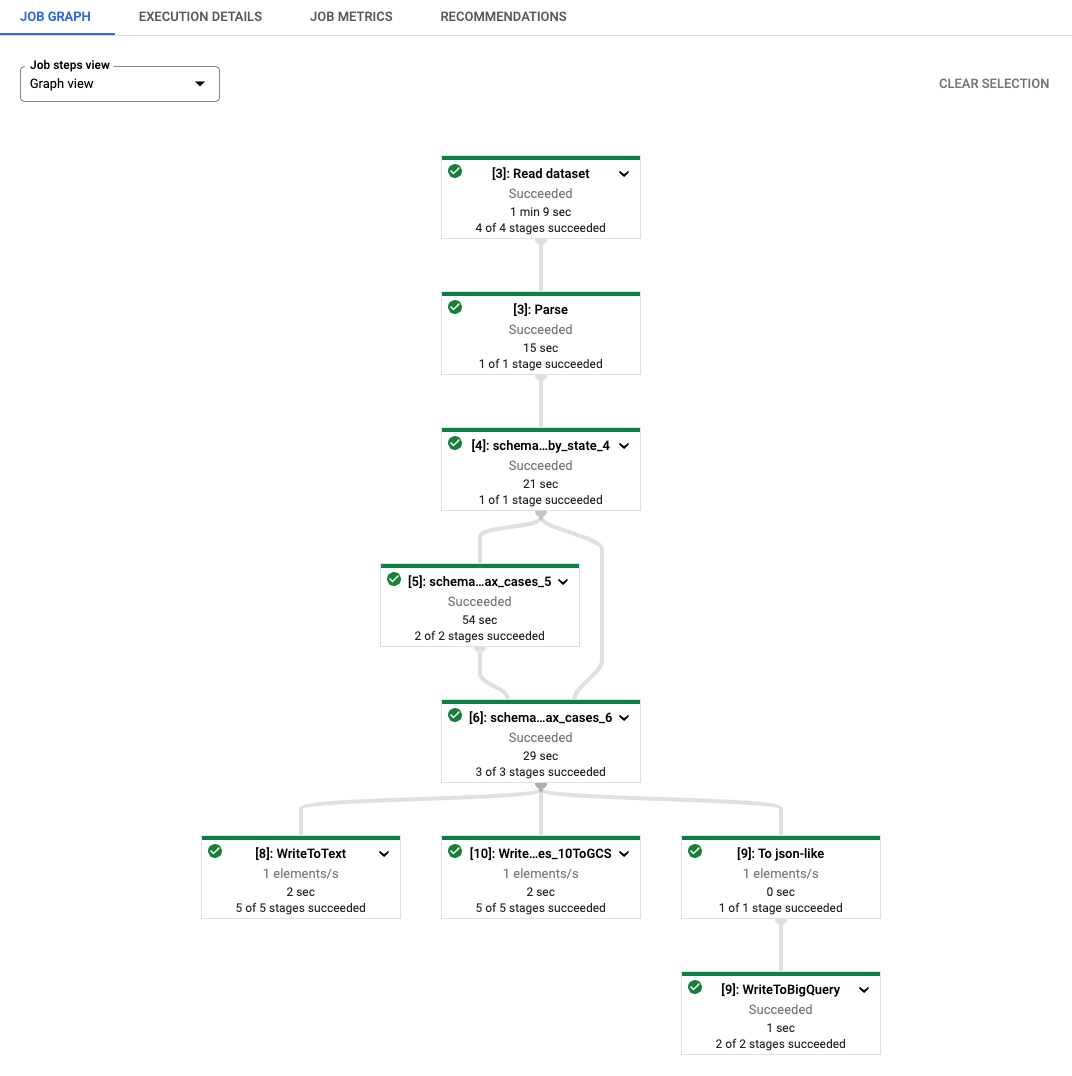
As you can see, each transform name of the generated Dataflow job is prefixed
with a string [number]: . This is to distinguish re-executed codes in
notebooks because Beam requires each transform to have a distinct name. Under
the hood, the beam_sql magic also stages your schema information to Dataflow,
so you might see transforms named as schema_loaded_beam_sql_…. This is because
the NamedTuple defined in the notebook is likely in the __main__ scope and
Dataflow is not aware of them at all. To minimize user intervention and avoid
pickling the whole main session (and it’s infeasible to pickle the main session
when it contains unpickle-able attributes), the beam_sql magic optimizes the
staging process by serializing your schemas, staging them to Dataflow, and then
deserialize/load them for job execution.
Once the job succeeds, the result of the output PCollection would be written to
places instructed by your I/O transforms. Note: running beam_sql on
Dataflow generates a one-shot job and it’s not interactive.
A simple inspection of the data from the default output location:
!gsutil cat 'gs://ningk-so-test/bq/staging/data_with_max_cases*'

The text file with encoded binary data written by your WriteToText:
!gsutil cat 'gs://ningk-so-test/bq/encoded_max_data*'

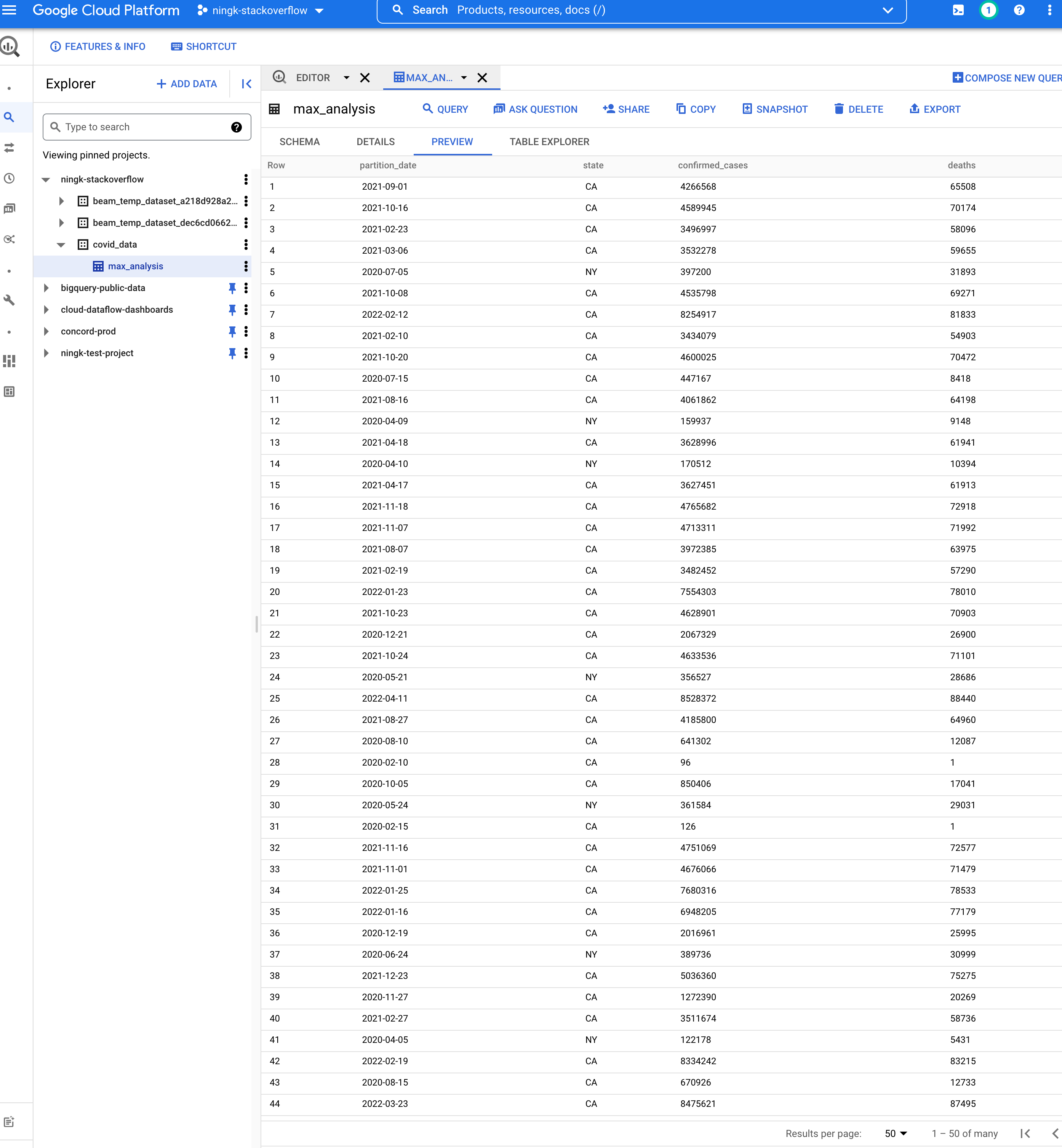
The table YOUR-PROJECT:covid_data.max_analysis created by your
WriteToBigQuery:
Run on other OSS runners directly with the beam_sql magic
On the day this blog is posted, the beam_sql magic only supports DirectRunner
(interactive) and DataflowRunner (one-shot). It’s a simple wrapper on top of
the SqlTransform with interactive input widgets implemented by
ipywidgets. You can implement
your own runner support or utilities by following the
instructions.
Additionally, support for other OSS runners are WIP, for example,
support using FlinkRunner with the beam_sql magic.
Conclusions
The beam_sql magic and Apache Beam Notebooks combined is a convenient tool for
you to learn Beam SQL and mix Beam SQL into prototyping and productionizing (
e.g., to Dataflow) your Beam pipelines with minimum setups.
For more details about the Beam SQL syntax, check out the Beam Calcite SQL compatibility and the Apache Calcite SQL syntax.



